Autoensamblaje de cápsides virales
Teoría y Simulación
Abstract
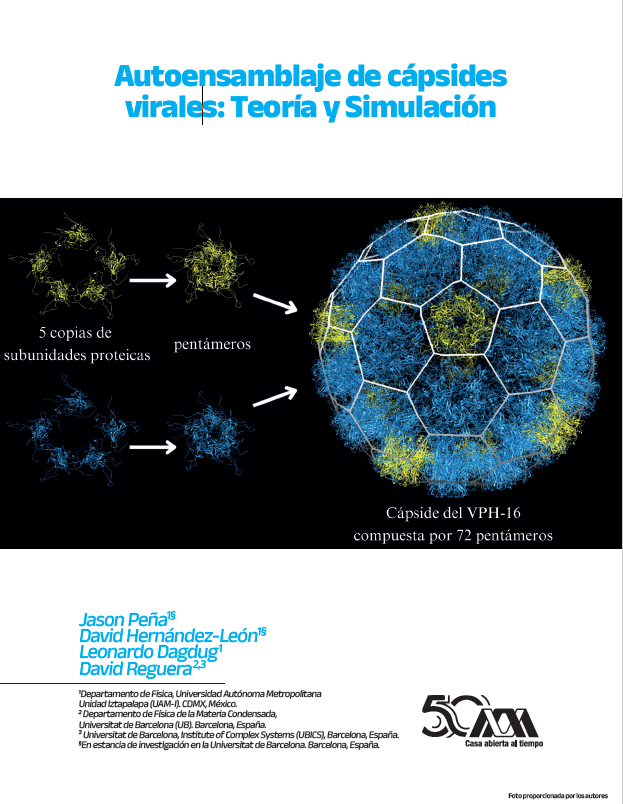
Viruses, despite their diminutive size, undoubtedly play an important role in human life and history. Nowadays, virology constitutes a prominent and interdisciplinary research field with diverse applications within disciplines such as biology, biomedicine, and public health. The fascinating and uncommon properties of viruses, as well as the intricate mechanisms they follow in the replication cycle, have captured the attention of physics and the basic sciences, giving rise to the emerging and promising field of physical virology. This article addresses the collaborative efforts of the Department of Condensed Matter Physics at Universidad de Barcelona (UB) and the Physics Department at Universidad Autónoma Metropolitana Iztapalapa (UAM-I), to unravel, using statistical physics methodologies, one of the fundamental steps in the viral replication cycle: the assembly of the protein capsid that protects viral genetic material. It provides an overview of the theoretical framework employed to model Viral Capsid Self-Assembly (VCSA), rooted in Classical Nucleation Theory (CNT), focusing on viruses whose capsids exhibit spherical symmetry. This framework incorporates key physical factors, such as protein concentration, temperature, and edge energy, which are critical determinants of the assembly process. Within this context, the role of bending energy is analyzed, as it can potentially alter assembly pathways and open the door to novel strategies to interfere with virus formation. This article further explores the utility of Molecular Dynamics (MD) simulations as a powerful computational tool to characterize the interactions between capsid proteins of specific viruses, such as the Human Papillomavirus (HPV). By analyzing these simulations, valuable insights can be obtained into the mechanisms of capsid assembly and disassembly, which are fundamental for a deeper understanding of viruses and the development of innovative antiviral strategies.
Downloads
References
Buzón P., Maity S. y Roos W. H.,Physical virology: From virus self-assembly to particle mechanics, Wiley Interdiscip Rev Nanomed Nanobiotechnol, 12(4):e1613, 2020.
Dimitrov D., Virus entry: molecular mechanisms and biomedical applications, Nat Rev Microbiol., 2(2):109-22, 2004.
Hagan M. y Grason G., Equilibrium mechanisms of self-limiting assembly. Rev Mod Phys. 93(2):025008, 2021.
Luque A. y Reguera D., Theoretical Studies on Assembly, Physical Stability and Dynamics of Viruses, en Structure and Physics of Viruses: An Integrated Textbook, ed. por Mateu M. G., Springer Netherlands, Dordrecht, pp. 553-595, 2013.
Mateu M. G., The Structural Basis of Virus Function, en Structure and Physics of Viruses: An Integrated Textbook, ed. por Mateu M. G., Springer Netherlands, Dordrecht, pp. 3-51, 2013.
Mendoza, C. I. y Reguera, D., Shape selection and mis-assembly in viral capsid formation by elastic frustration. Elife, 9, e52525, 2020.
Mejía-Méndez, J. L., Vazquez-Duhalt, R., Hernández, L. R., Sánchez-Arreola, E., y Bach, H., Virus-like particles: fundamentals and biomedical applications, International journal of molecular sciences, 23(15), 8579, 2022.
Smith, K. M., Viruses, Cambridge University Press, 1963.
Shirbaghaee, Z. y Bolhassani, A., Different applications of virus-like particles in biology and medicine: Vaccination and delivery systems. Biopolymers, 105: 113- 132, 2016.
Vehkamäki, H, Classical Nucleation Theory in Multicomponent Systems, Springer Berlin, Heidelberg, 2006.
Zandi R, van der Schoot P, Reguera D, Kegel W, Reiss H. Classical nucleation theory of virus capsids. Biophys J., 90[6], pp.1939-48, 2006.